Quantum simulation offers a groundbreaking approach for scientists to delve into the depths of complex systems that are otherwise insurmountable by classical computers. This technology has opened up new possibilities in various fields, ranging from financial modeling to cybersecurity, pharmaceutical discoveries, AI, and machine learning. The ability to explore molecular vibronic spectra, a critical aspect of understanding molecular properties in molecular design and analysis, has long been a computational challenge for traditional supercomputers.
Although researchers are diligently working on quantum computers and algorithms to simulate molecular vibronic spectra, they have faced limitations in dealing with complex molecule structures, accuracy issues, and inherent noise. The recent breakthrough by engineering researchers at The Hong Kong Polytechnic University (PolyU) has set a new milestone in the field. They have successfully developed a quantum microprocessor chip capable of simulating the molecular spectroscopy of large and complex molecules, marking a significant advancement in quantum technology.
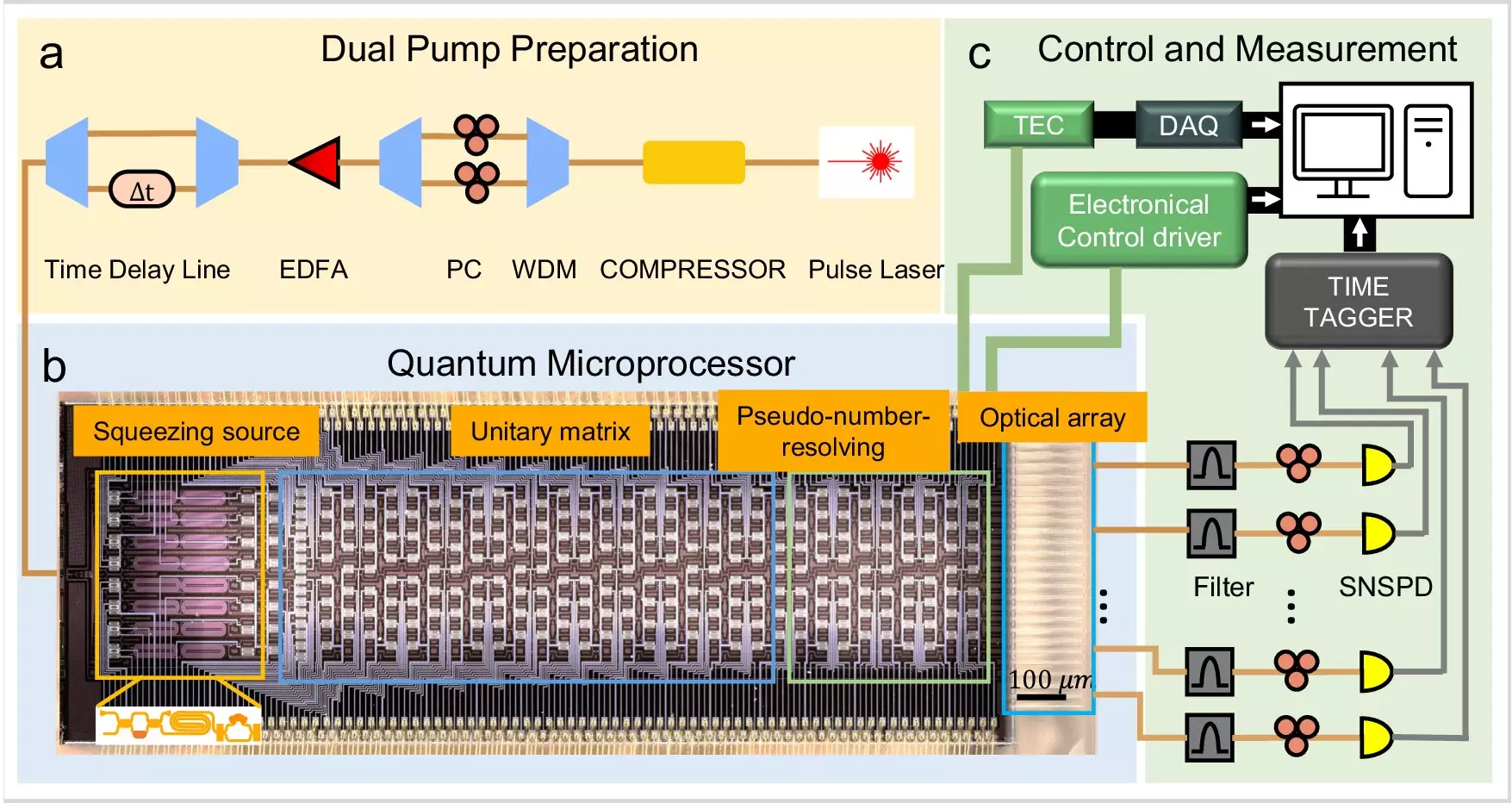
Accurate simulation of quantum effects such as superposition and entanglement is essential in capturing the nuances of molecular vibronic spectra. These effects are computationally intensive to model classically, making quantum simulation the preferred approach for tackling such complex problems. The research team led by Professor Liu Ai-Qun and Dr. Zhu Hui Hui has leveraged a large-scale photonic network with squeezed vacuum states to enable precise molecular spectroscopy simulation, as highlighted in their research paper published in Nature Communications.
The development of a 16-qubit quantum microprocessor chip integrated into a single chip opens up a myriad of possibilities for quantum computational applications that surpass the capabilities of classical computers. This cutting-edge technology lays the foundation for solving intricate quantum chemistry problems and conducting molecular simulations with unprecedented speed and accuracy. The researchers envision the application of quantum microprocessors in simulating large protein structures and optimizing molecular reactions to revolutionize the field of quantum chemistry.
The integration of quantum microprocessor chips in molecular spectroscopy has paved the way for significant advancements in scientific fields such as material science, chemistry, and condensed matter physics. These quantum technologies offer promising alternatives for quantum information processing and hold immense potential for practical applications in molecular docking, quantum machine learning, and graph classification. Professor Liu emphasized the real-world impact of quantum simulation technologies and expressed their commitment to scaling up the microprocessor for tackling more complex applications with societal and industrial benefits.
The successful development of a quantum microprocessor chip for molecular spectroscopy simulation represents a major breakthrough in quantum technology and its applications. The transformative impact of quantum simulation on complex systems, particularly in molecular spectroscopy, underscores the immense potential of quantum technologies in advancing scientific research and technological innovations. The research findings by PolyU’s engineering researchers have set the stage for unveiling new avenues in quantum computing and pushing the boundaries of what is achievable in the realm of molecular analysis and simulation.
Leave a Reply